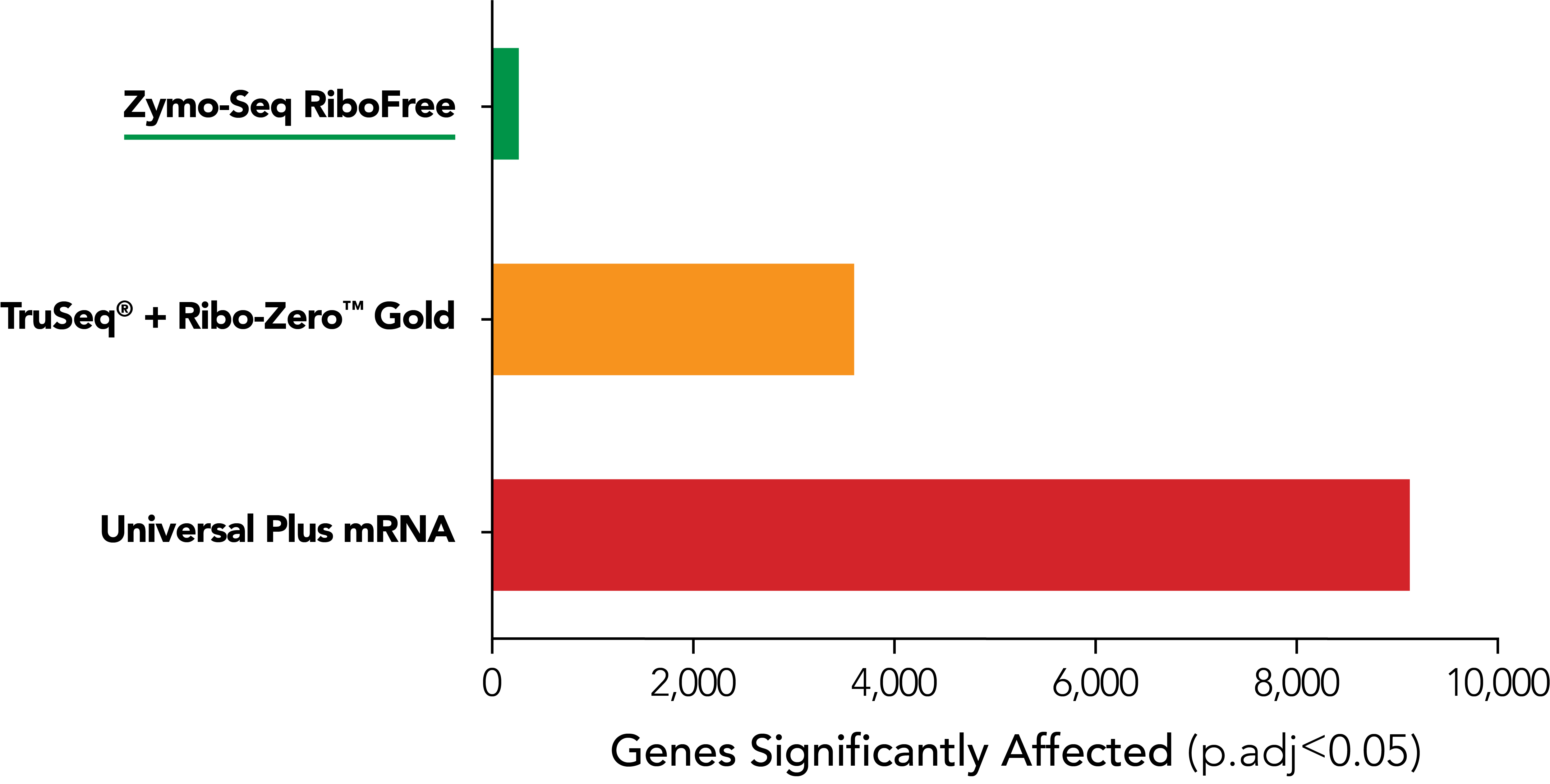Cat #: 11-643B
Zymo Research R3003 Zymo-Seq RiboFree Total RNA Library Kit, Zymo Research, 96 Preps/Unit


Cat #: 11-643B
Zymo Research R3003 Zymo-Seq RiboFree Total RNA Library Kit, Zymo Research, 96 Preps/Unit
Zymo Research
96 Preps/Unit
Brand: Zymo Research- The Fastest & Easiest Total RNA-Seq Library Kit: Prepare stranded, RiboFree libraries from total RNA in 3.5 hours
- Compatible with Any Sample: Probe-free technology depletes rRNA & Globin from any RNA source
- The Most Accurate: Eliminate bias from rRNA depletion

$8,125.85
Login To Access Your Institutional Price
Zymo Research
96 Preps/Unit
Brand: Zymo Research- The Fastest & Easiest Total RNA-Seq Library Kit: Prepare stranded, RiboFree libraries from total RNA in 3.5 hours
- Compatible with Any Sample: Probe-free technology depletes rRNA & Globin from any RNA source
- The Most Accurate: Eliminate bias from rRNA depletion
Zymo-Seq RiboFree Total RNA-Seq Library Kit is the fastest, easiest RNA-Seq library prep kit available to make stranded, total RNA libraries. The automation-friendly protocol minimizes hands-on time to generate sequencer-ready, stranded, indexed libraries depleted of rRNA and globin in as little as 3.5 hours. Depletion of ribosomal RNAs (which can comprise as much as 90% of a total RNA sample) is completely integrated into the workflow, using a novel, probe-free technology that removes rRNA, globin, or any other over-abundant, repetitive transcript from any sample type or species. RiboFree Universal Depletion is validated across biological kingdom and phyla, including human, rodent, plant, and various prokaryotes, as well as RNA from a wide range of sample types including cells, tissue, FFPE tissues, and whole blood, eliminating the need to select sample-specific modules for different sample types. Just one total RNA-Seq library prep kit to deplete them all. Because the RiboFree Universal Depletion is probe-free, researchers using the Zymo-Seq RiboFree Total RNA Library Kit will avoid transcriptome profiling bias found in other methods. High concentrations of rRNA-binding oligonucleotides used in probe-based approaches also bind to a significant number of non-ribosomal targets. As much as 20% of the 20,000 protein-coding genes in the human genome are significantly biased by these methods. In contrast, the probe-free depletion developed by Zymo Research uses only abundant pre-existing transcripts for mismatch-free enzymatic removal of rRNA and globin, resulting in minimal off-target effects as low as 1.8% in all protein-coding transcripts. This total RNA-Seq library prep kit’s protocol is fragmentation-free and works with up to 2 ug of undepleted total RNA, down to 50 ng of undepleted total RNA, or as little as 5 ng of ribosome-depleted RNA samples. Pre-mixed reagents are ready-to-use at every step, eliminating the need for manual master-mix preparation. Everything needed to make sequencing-ready libraries, including indexing primers, SPRI beads, and magnetic bead stand are included in the kit. It’s RNA-Seq made simple.
Enjoy our products? Leave a review and let us know.
Sample To Sequencer In A Single Day |
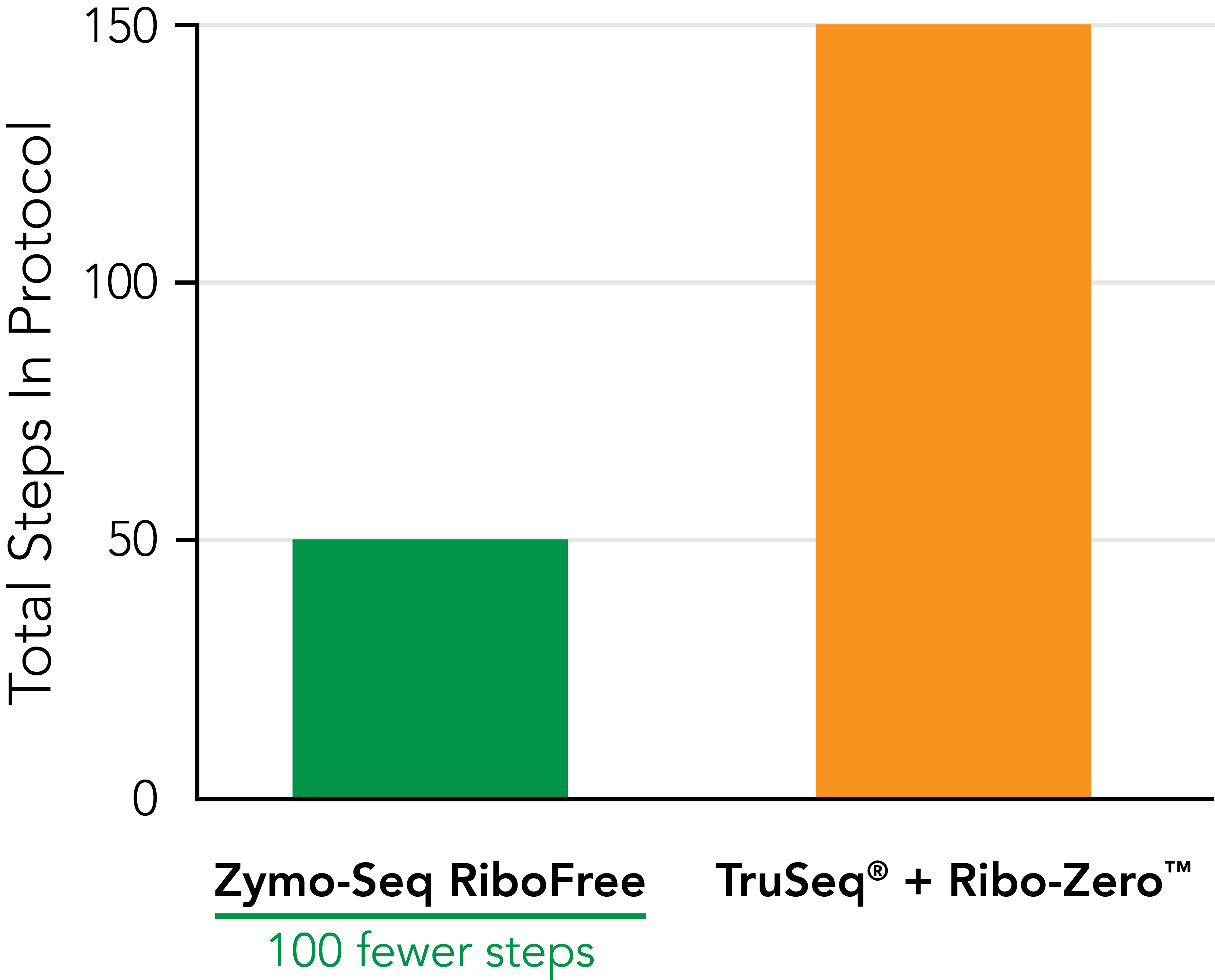
Significantly Reduce Hands‑on Time 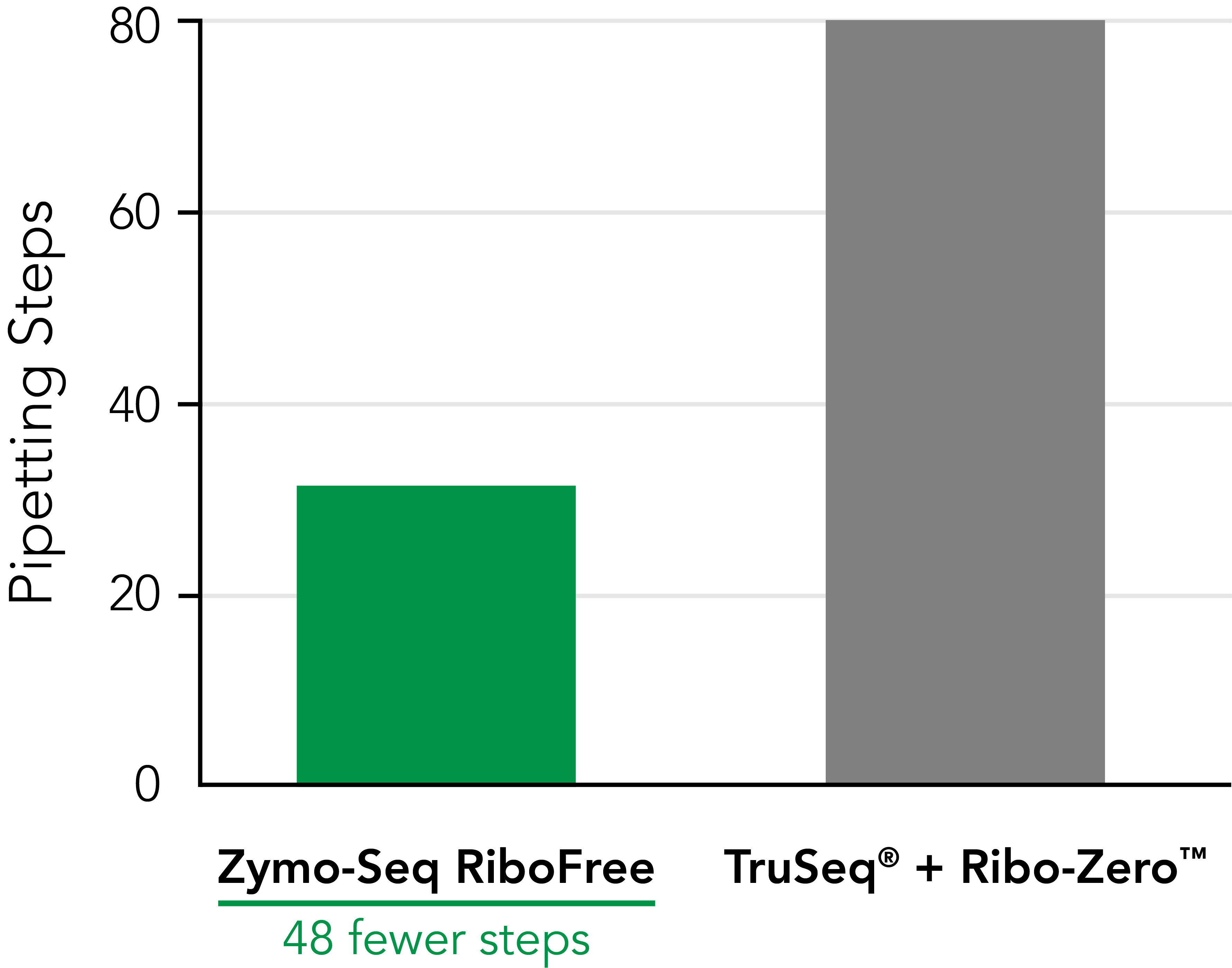 Zymo-Seq RiboFree Total RNA Library Kit significantly reduces pipetting steps compared to TruSeq® Ribo-ZeroTM Gold. An easy-to-follow protocol allows users to go from RNA samples to the sequencer in a single day. |
Probe-free rRNA Depletion
Compatible With Any Organism
Use One Kit For Any Sample Type
Make Any Library RiboFree
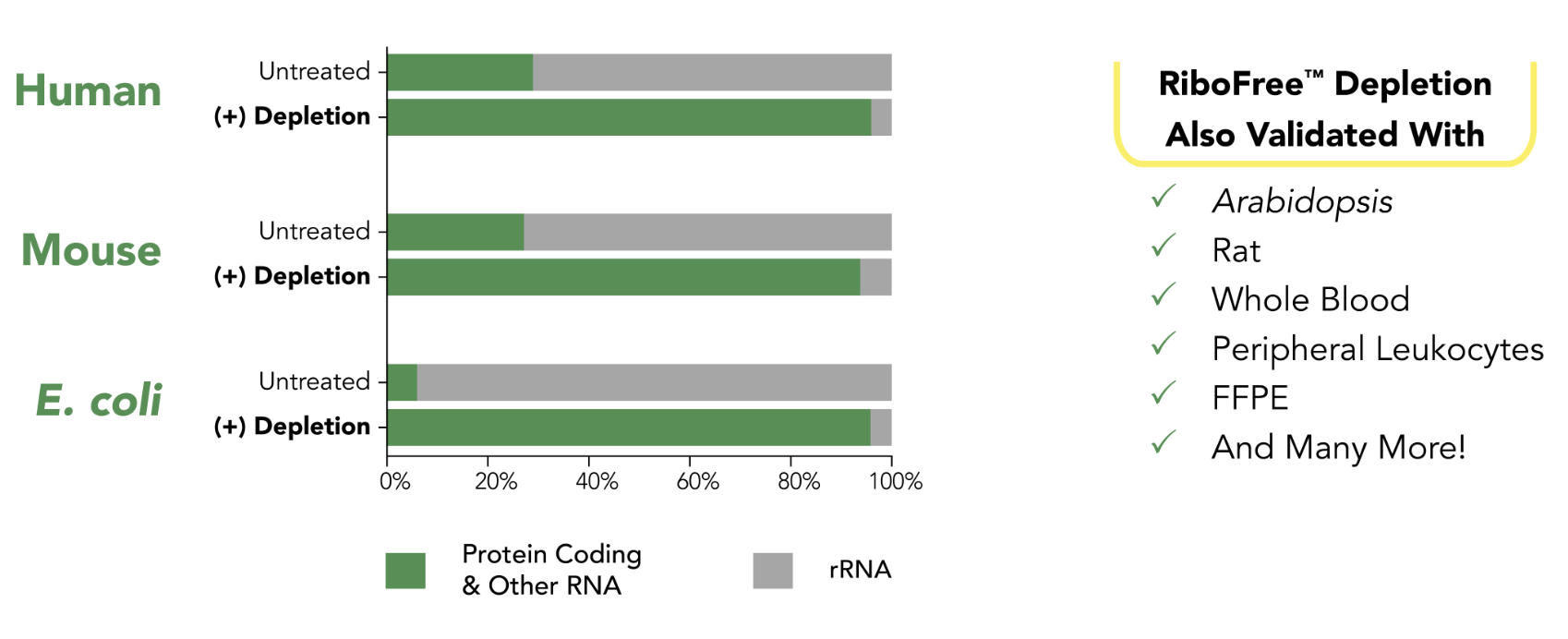
RiboFree depletion will remove rRNA from any sample type. Paired-end sequencing was performed on stranded total RNA-Seq libraries containing ERCC Spike-In Mix 1 (Life Technologies), both with and without RiboFree depletion. Read pairs were aligned to their respective genomes using the STAR aligner. Read classes were defined using a combination of Ensembl GTF gene biotypes and RepBase repeat masker annotations. Number of reads overlapping each annotation class were divided by total reads in that library to calculate percent reads of each annotation class.
Probe-free. Bias‑free. RiboFree.
The Most Accurate rRNA Depletion
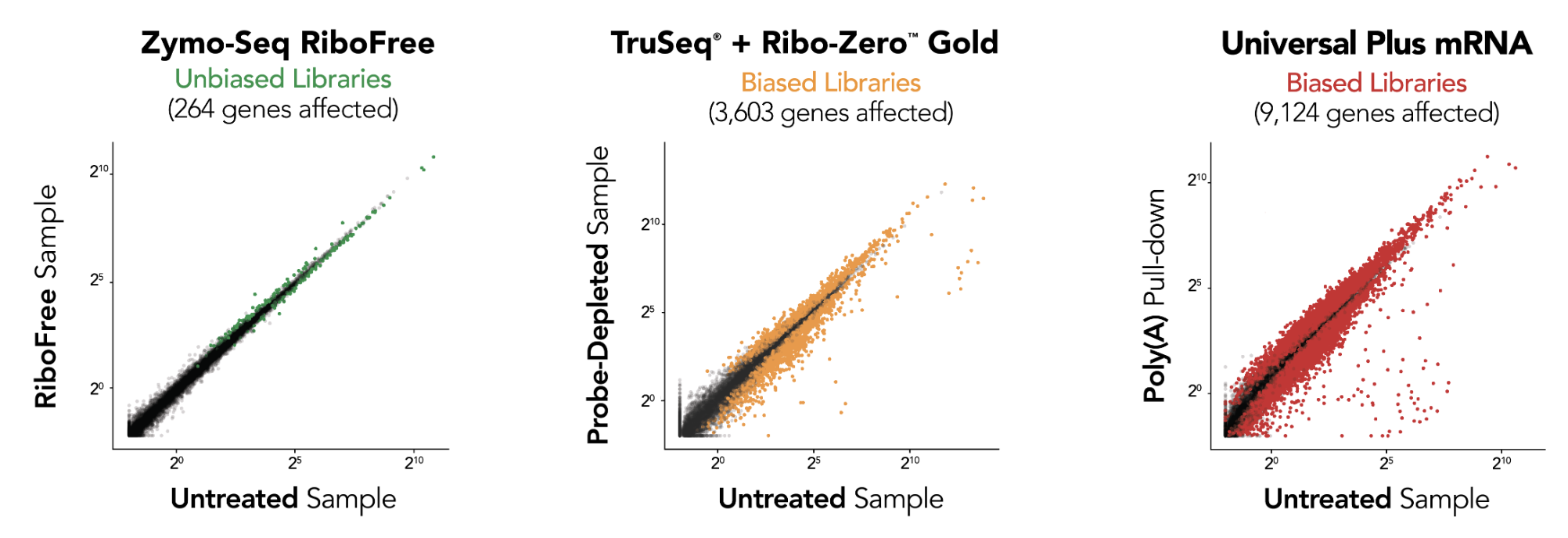
RiboFree depletion maintains native expression profiles unlike Supplier i [probe-based Ribo-ZeroTM Gold] and Supplier NG [poly(A) enrichment]. Paired-end sequencing was performed on libraries prepared from Universal Human Reference RNA (Invitrogen) containing ERCC Spike-In Mix 1 (Life Technologies), both with and without rRNA removal. Libraries were sequenced to a depth of ∼35 million reads per library, and read pairs were aligned to the hg38 human genome using the STAR aligner. The DESeq2 package was used to apply "apeglm" as a log-fold-change shrinkage correlation and to determine which of the 20,004 protein coding genes and ERCC Spike-In transcripts were significantly affected (p.adj < 0.05) by rRNA removal. Significantly affected transcripts are represented as colored dots in the scatterplots.
| Zymo Research | Supplier I | Supplier NG | |
|---|---|---|---|
| Stranded | |||
| Total time (including depletion) | 3.5 hours | >9.5 hours | >8.5 hours |
| Total steps in protocol | 51 | 151 | 127 |
| One kit. Any organism. | |||
| All reagents included from input to indexing |