Cat #: 11-602B
Zymo Research D6306 Microbial Community DNA Standard, ZymoBIOMICS™, 2000ng/Unit










Cat #: 11-602B
Zymo Research D6306 Microbial Community DNA Standard, ZymoBIOMICS™, 2000ng/Unit
ZymoBIOMICS™
2000ng/Unit
Brand: Zymo Research- DNA of a mock microbial community of well-defined composition
- Ideal for the validation and optimization of microbiomics or metagenomcis workflows
- Ideal as quality control of microbiome profiling and metagenomic analyses
- Source: a mixture of genomic DNA of ten microbial strains.
- Storage solution: 10 mM Tris-HCl and 0.1 mM EDTA, pH 8.0
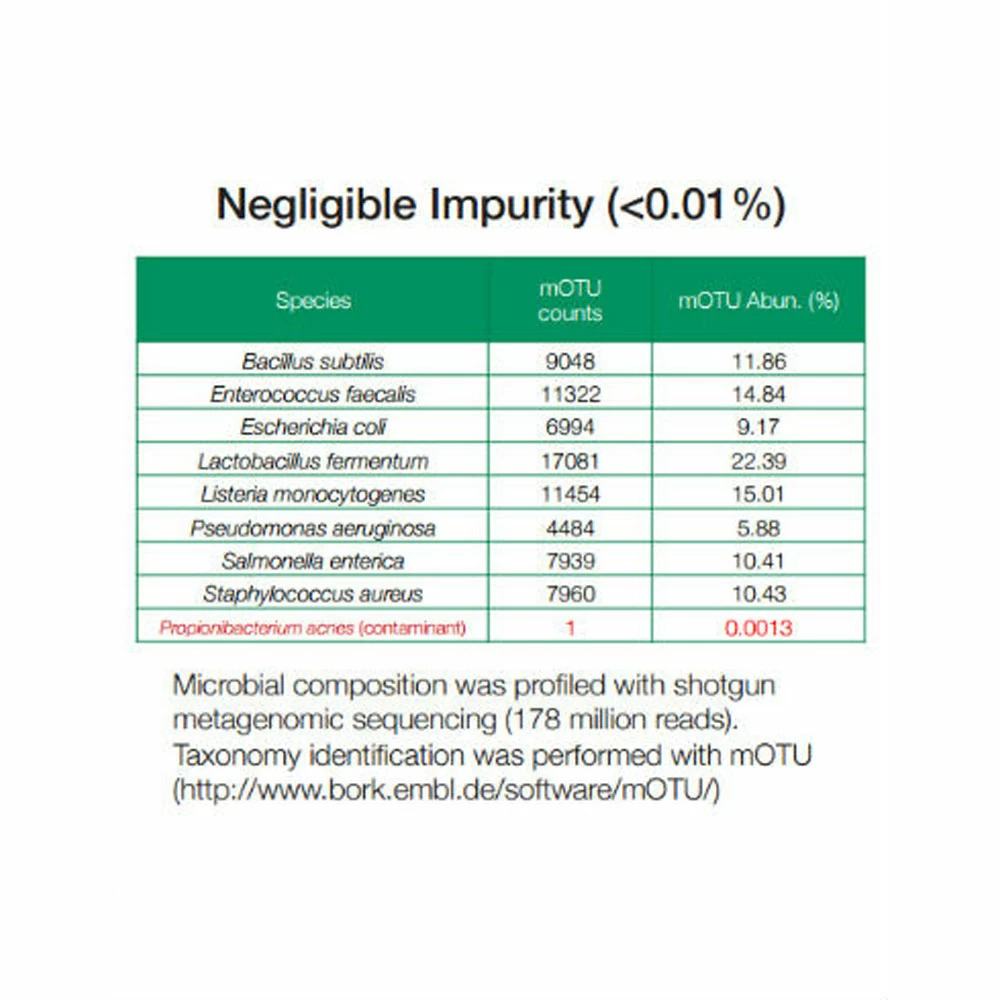
- Impurity level: < 0.01% foreign microbial DNA
ZymoBIOMICS™
2000ng/Unit
Brand: Zymo Research- DNA of a mock microbial community of well-defined composition
- Ideal for the validation and optimization of microbiomics or metagenomcis workflows
- Ideal as quality control of microbiome profiling and metagenomic analyses
- Source: a mixture of genomic DNA of ten microbial strains.
- Storage solution: 10 mM Tris-HCl and 0.1 mM EDTA, pH 8.0
- Impurity level: < 0.01% foreign microbial DNA
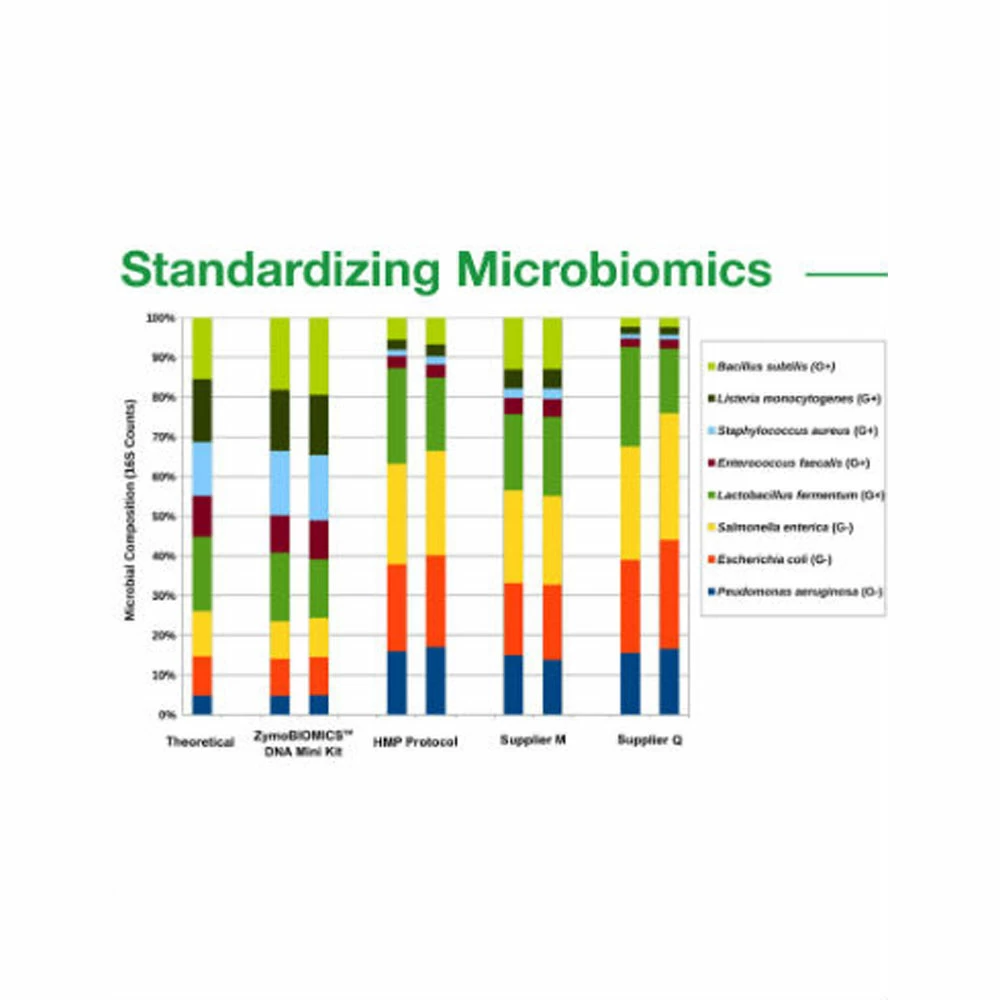
ZymoBIOMICS™ Microbial Community DNA Standard is a mixture of genomic DNA extracted from pure cultures of eight bacterial and two fungal strains. Genomic DNA from each pure culture was extracted and quantified before mixing. The GC content of the containing genomes covers a range from 15% to 85%. The microbial standard is accurately characterized and contains negligible impurities (< 0.01%). This enables it to expose artifacts, errors, and bias in microbiomics or metagenomics workflows. The DNA Standard is ideal for assessing biases and errors in processes of library preparation, sequencing, and bioinformatics analyses. It serves perfectly as a microbial standard for benchmarking the performance of microbiomics or metagenomics analyses and to serve as a control in inter-lab studies. This standard is also ideal to help users construct and optimize workflows, e.g. controlling PCR chimera rate and noise in the library preparation of 16S rRNA gene targeted sequencing, and assessing GC bias in sequencing coverage of shotgun metagenomic sequencing).
Details regarding the ten microbial strains (including species name, genome size, ploidy, average GC content, 16S/18S copy number, phylogeny) can be found in Table 2 on the “more information” link below. The 16S/18S rRNA sequences (fasta format) and genomes (fasta format) of these strains are available upon request. Feel free to contact us if you need help analyzing the sequencing data generated from this standard.
Enjoy our products? Leave a review and let us know.


